Research
These are some of the things we're now doing.
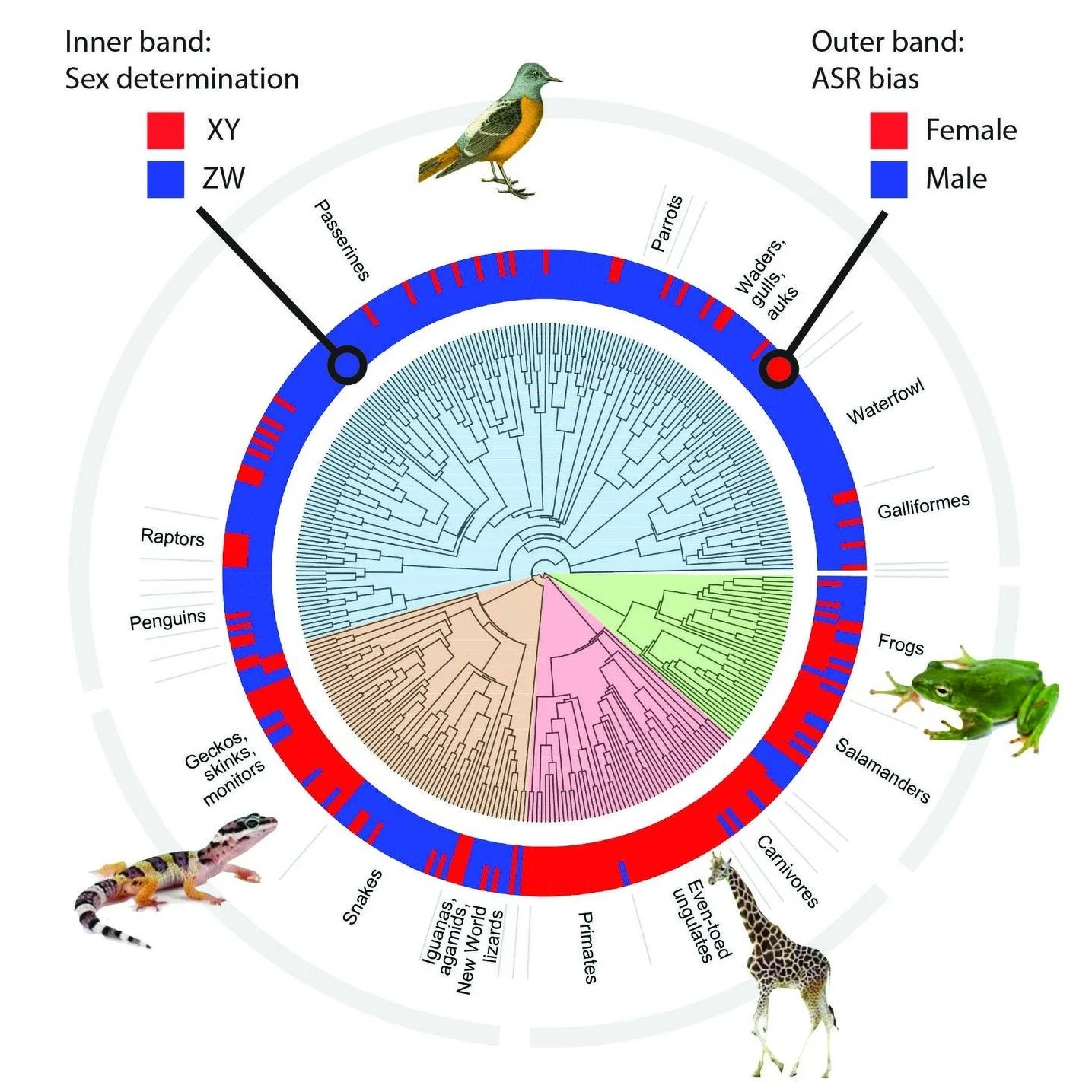
Sex chromosomes and sex determination
Sex chromosomes are a genomic hotspot for evolutionary change in many groups of animals and plants. Why do some species have XY, some ZW, and others polygenic sex determination? Our models show that sex antagonistic selection, often driven by sexual selection, can play a key role. We are testing that and other hypotheses in a long-term collaboration with Dr. Katie Peichel (University of Bern). Together we are analyzing genomic data from several species of stickleback fishes to understand the forces driving the very rapid evolution of their sex chromosomes.
Featured papers:
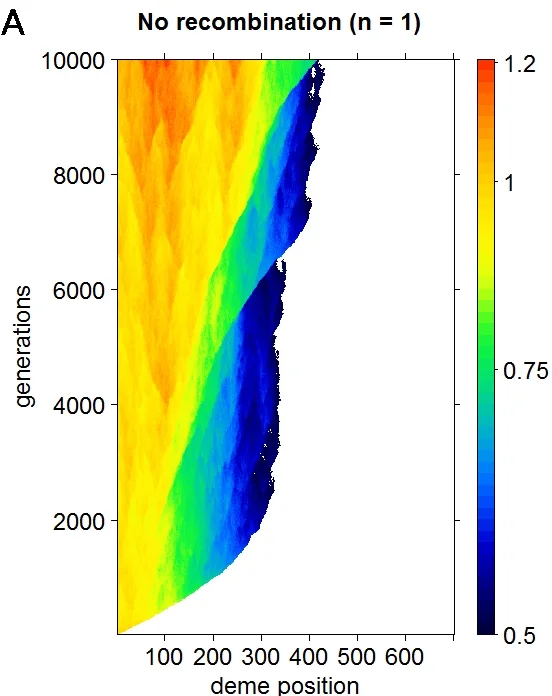
Genome evolution
Why are chromosomes so evolutionarily dynamic? In many taxa, chromosome rearrangements (such as inversions) are highly polymorphic within species and differ dramatically between species. Our models show that local adaptation resulting from ecological forces can be a powerful driver of genome architecture. We are quantifying the evolutionary forces acting on chromosome rearrangements using genetic and genomic data.
Featured papers:
Species ranges
Why don’t species adapt to marginal habitats and expand their ranges outwards? We work with models to explore mechanisms that put evolutionary limits on ranges. These include gene flow, biotic interactions, and genetic constraints.
Featured papers:
Sexual selection and speciation
We use models to learn how sexual selection can drive speciation, and we develop new statistical methods to quantify the evolutionary forces at work. We are also interested in the rules that animals use to choose their mates.
Featured papers:
Quantitative genetics
Traits like growth trajectories and reaction norms are “function-valued”: they are described by a continuous curve rather than a single measurement. We work with models to predict how these traits evolve, and develop statistical methods to estimate patterns of genetic variation for them. Another area of interest is to determine how patterns of genetic variation for multiple traits constrain evolution. Here we ultimately aim to estimate the evolutionary dimensionality of organisms and learn what limits rates of adaptation.
Featured papers:
Software
Recombining sex chromosomes in frogs
A PDF of the paper is here (click on this link).
The code is here (click on this link).
Selection on a mosquito inversion
A PDF of the paper is here (click on this link).
The code is here (click on this link).